- This topic has 17 replies, 3 voices, and was last updated February 20 by .
-
Topic
-
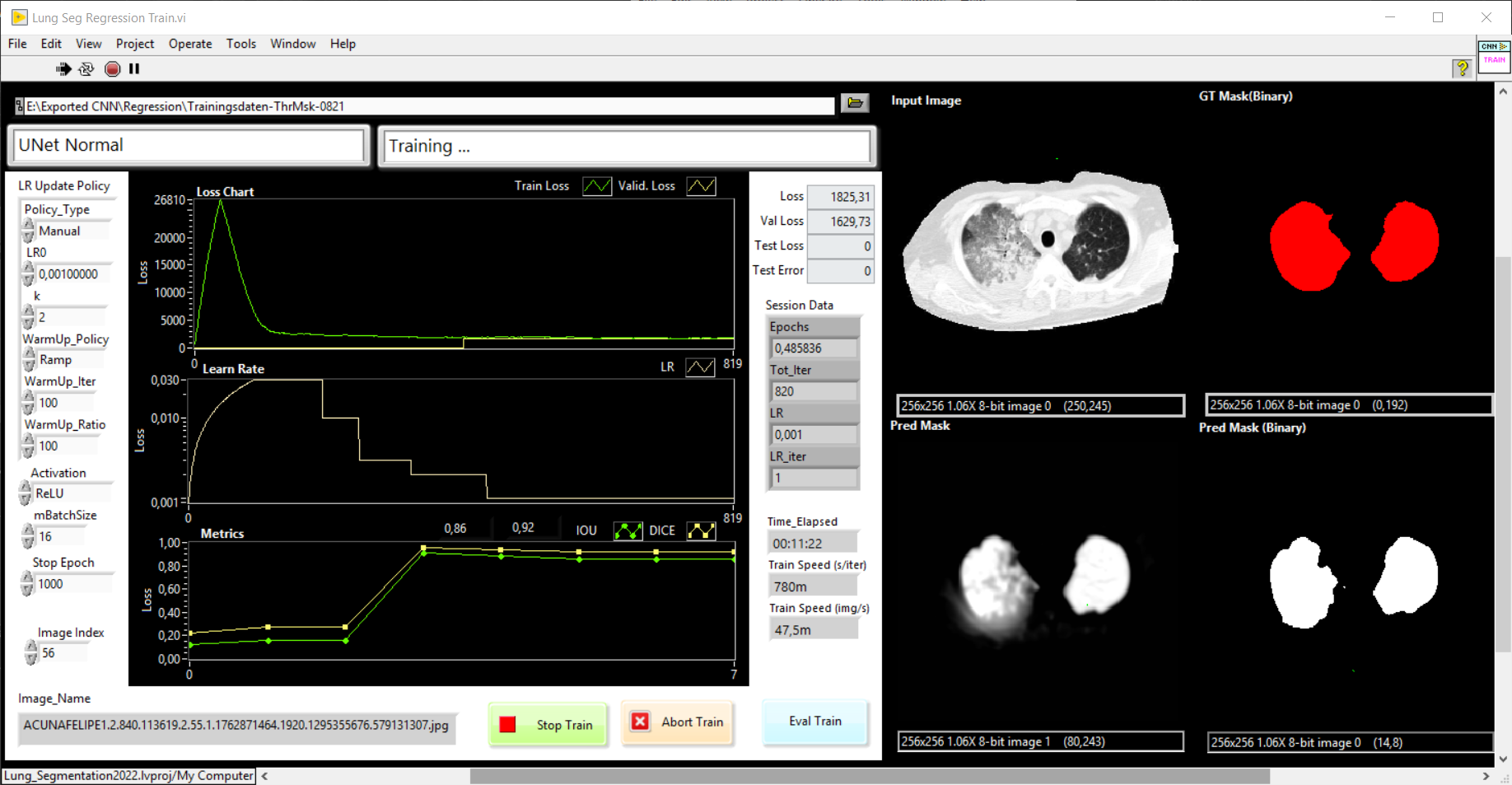
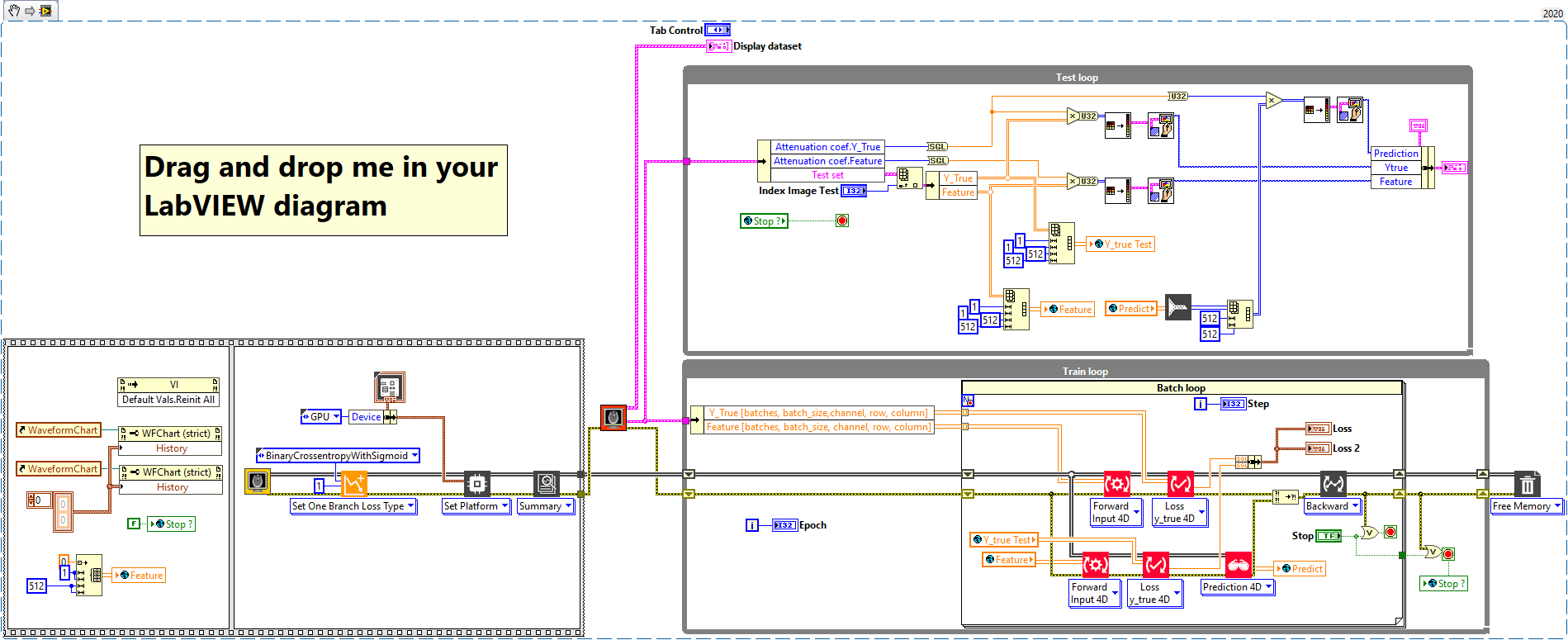
Our goal is the development of a UNet with Haibal for the automatic segmentation of lungs in computed tomographic (CT) images. First as a 2D UNet to segment the lung into individual CT slices. The image data are individual slices with a slice thickness of 0.6 mm to 5 mm. The complete lung can be reconstructed and rendered from a complete stack of images (with a slice thickness of 5 mm, that is about 50 – 60 slices). That’s why we would like to program a 3D UNet with Haibal (which is not possible with DeepLTK) in order to use the full information of the volume for segmentation. Predicted lung masks and ground truth (manually created masks) should be compared with the intersection over union (IoU) metric. Also known as the Jaccard index.
How can we realize a UNet with Haibal?
Previous work on Ngene’s Deep Learning Toolkit
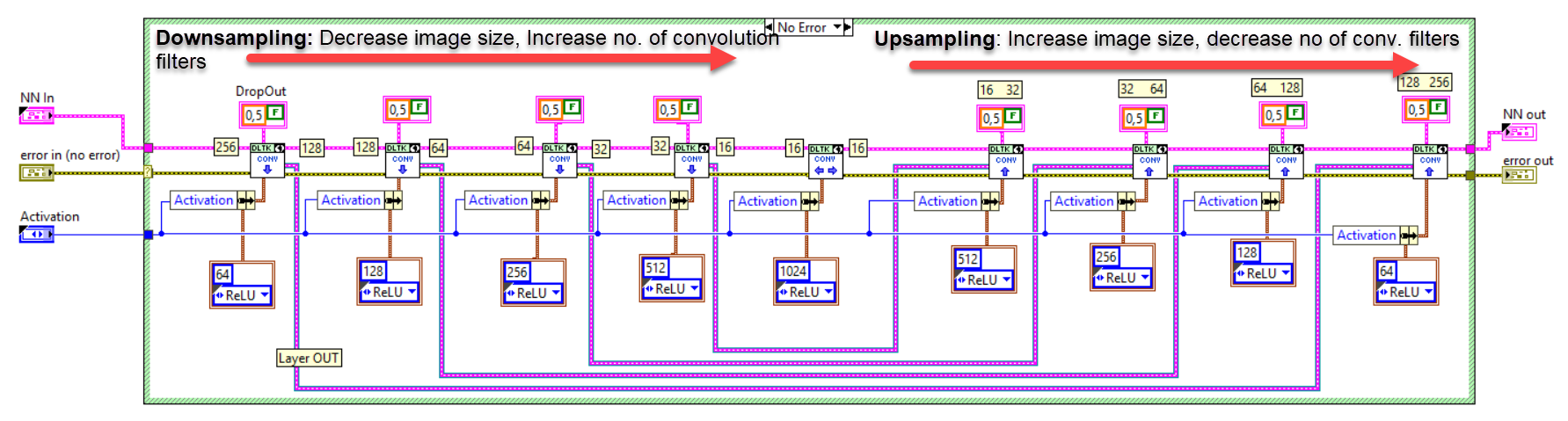
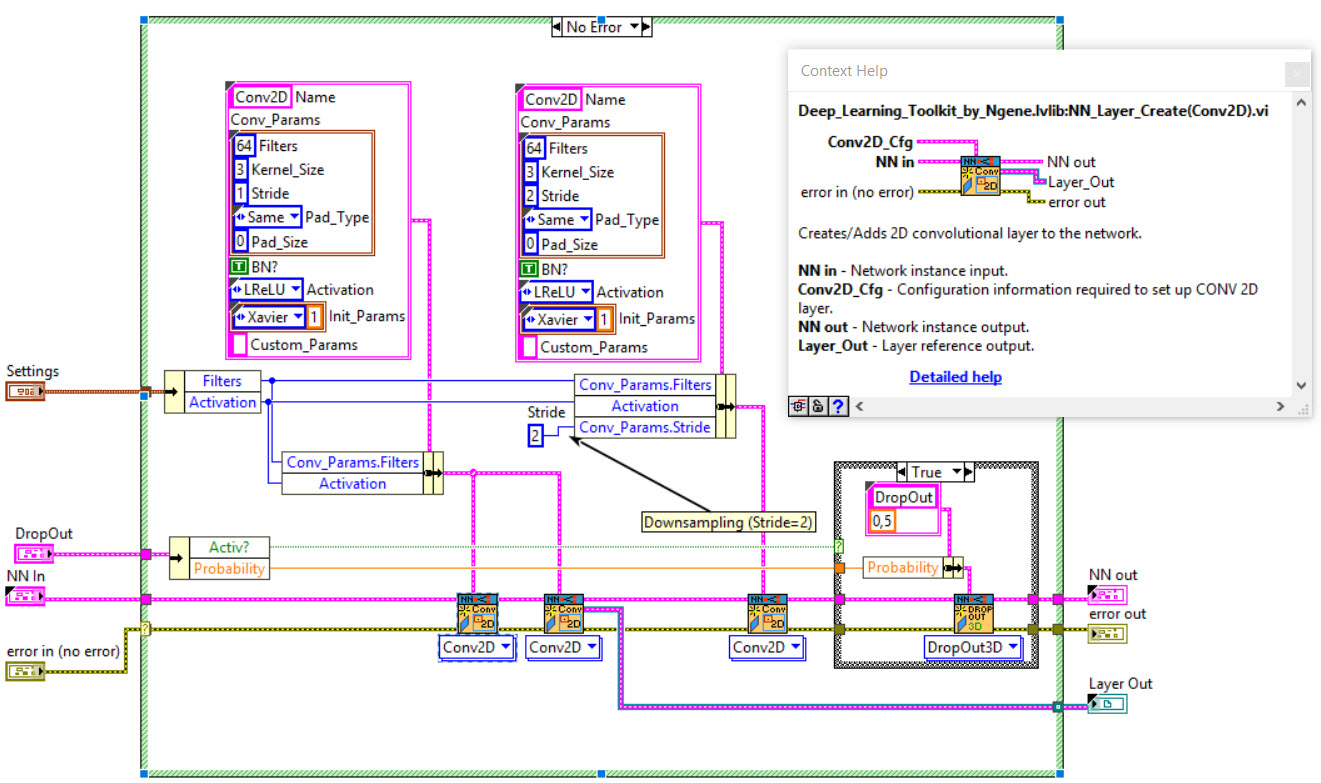
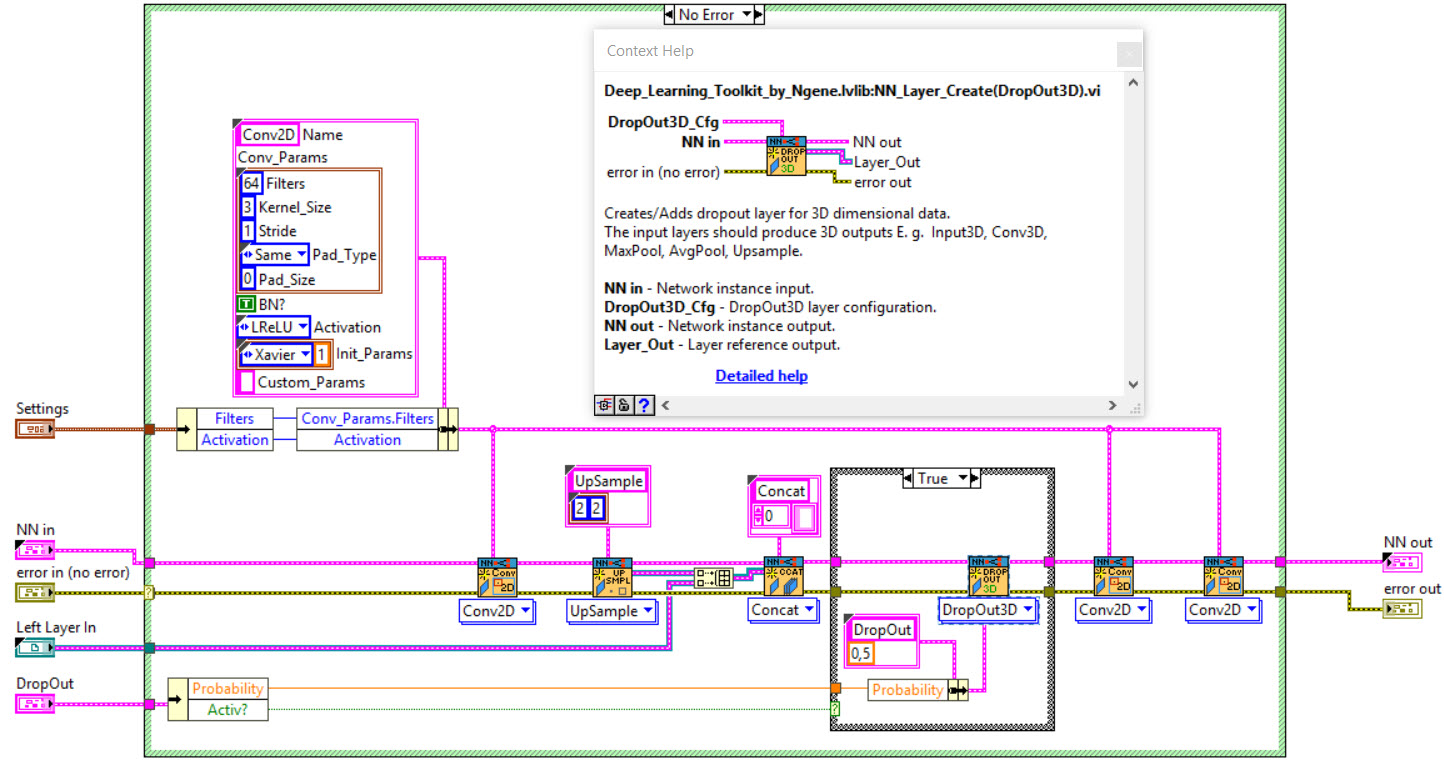
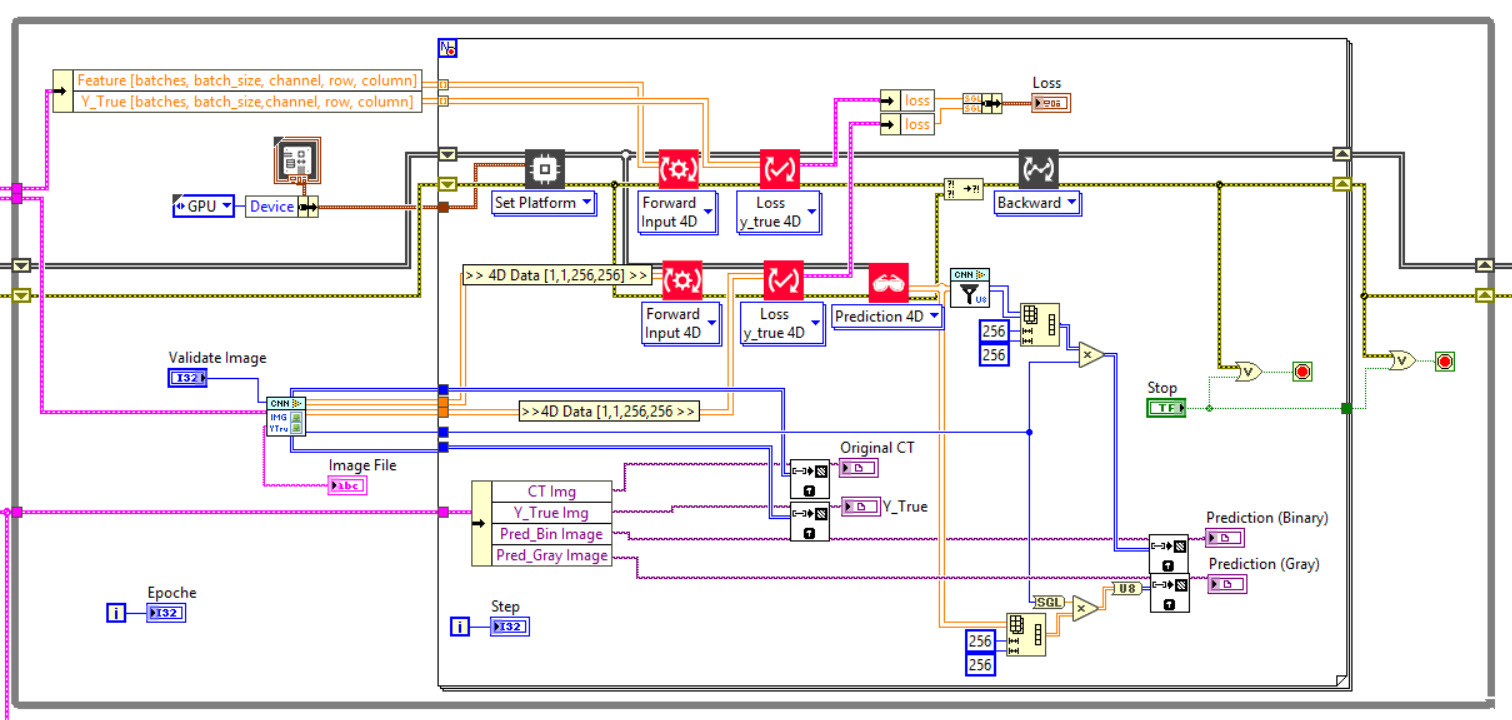
The network we used is based on the U-Net architecture. The U-Net was programmed with the graphical programming language LabVIEW , with which we had many years of experience in the development of software for image analysis, and the Deep Learning Toolkit for LabVIEW (Ngene, Armenia).The unique concept of U-Net is that it is able to generate a new, altered image as the output from an input image, after appropriate processing. This is very useful for generating segmentation images. The U-Net is a so-called fully convolutional network. Our U-Net programmed with LabVIEW is shown in Figure 1. The architecture has a symmetric “U” shape and consists of two major parts: a contraction path (left side) and an expansion path (right side). The path follows the typical architecture of a convolution neural network. It consists of the repeated application of two convolution layers, each layer with batch normalization, followed by an activation function. In all convolution layers we use a filter kernel size of 3 × 3 pixels. For each convolution we used the so called “SAME” padding type, which means there is automatically enough padding that the output image of the convolution layer has the same dimensions as the input image. For downsampling we chose a stride of 2 to halve the size of the input image. For upsampling we use an upsampling layer. This layer increases the dimensionality (rows and columns) of output feature maps by doubling the values (stride = 2).
The DeepLTK still has a number of limitations: IOU and Dice coefficient are the only metrics so far. More will be added in the next releases. Shape quality performance metrics like ASSD or BF-Score are not yet supported. The only optimization algorithms are SGD and Adam. Further algorithms such as Adagrad, AdaDelta, RMSProp, Nesterov … are being developed for the next releases. A 3D semantic segmentation architecture is still not possible with the DeepLTK.
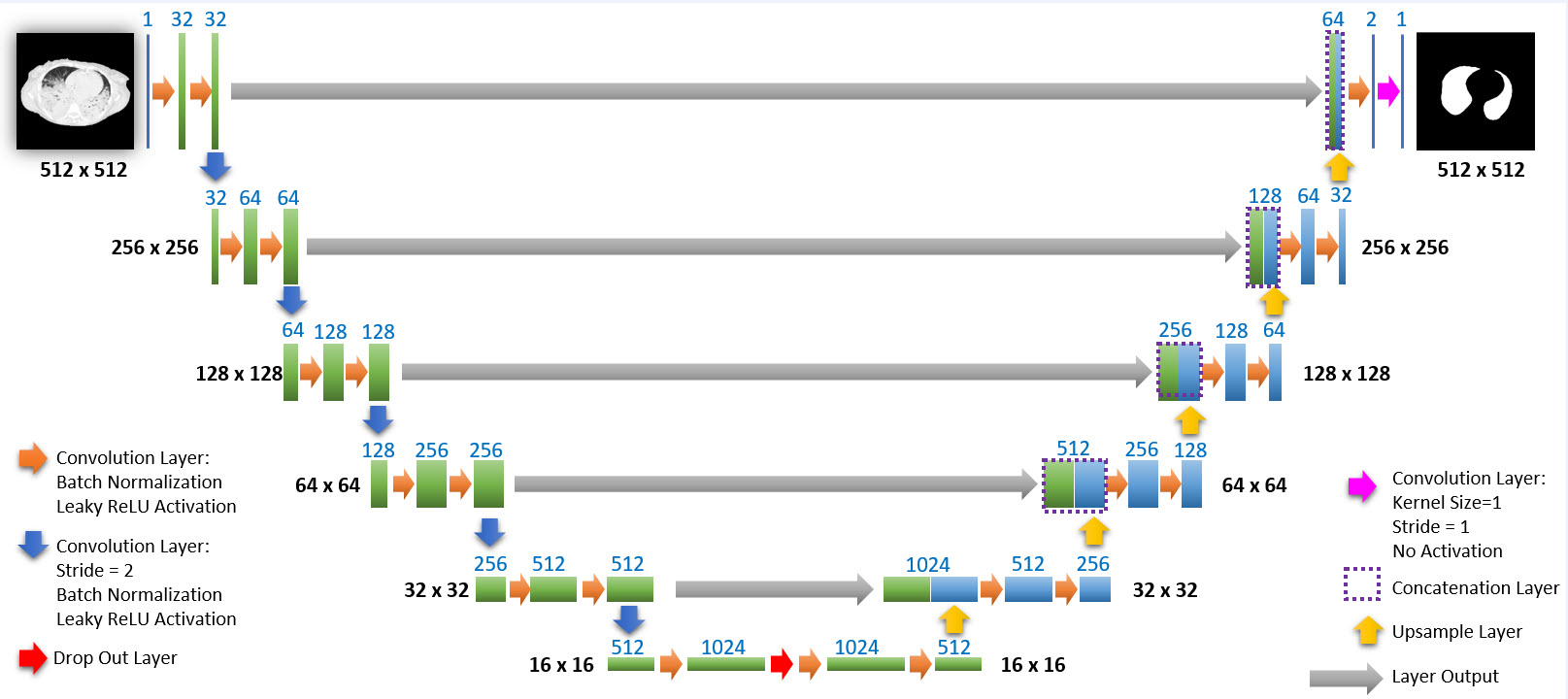
Figure 1: Our used U-Net architecture. Each green or blue box corresponds to a multi-channel feature map. The number of channels is shown above the box. The specifications 512 × 512 to 16 × 16 (in the lowest resolution) show the x, y dimensions in pixels of the input and output images (or feature maps).
References:
- You must be logged in to reply to this topic.