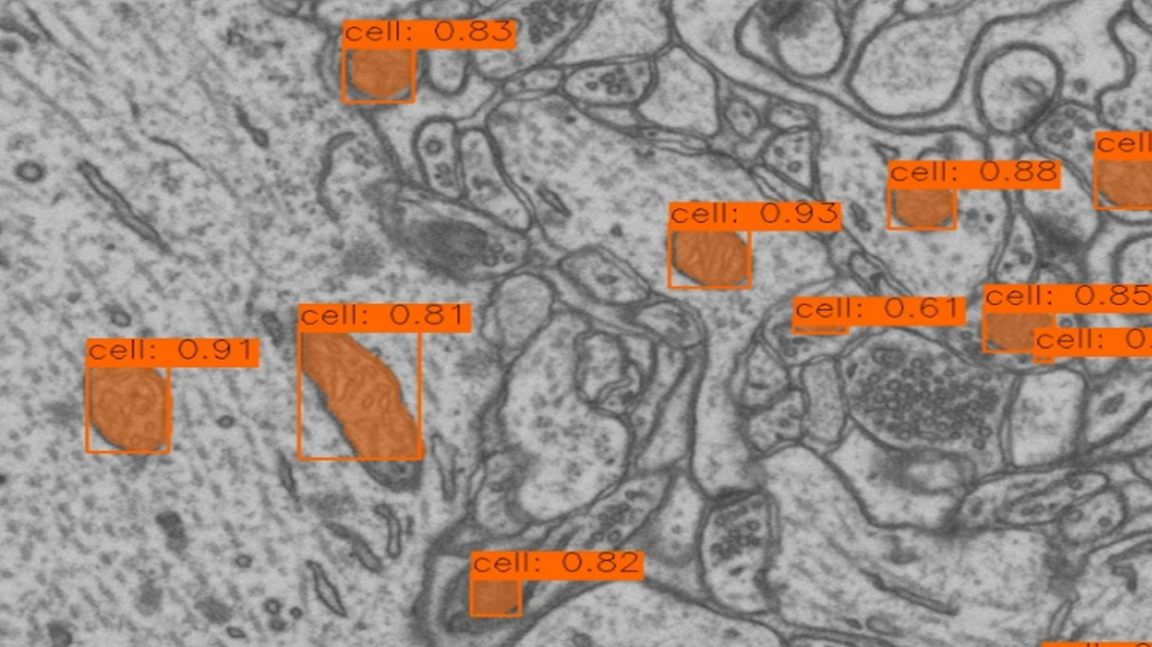
In the field of neuroscience research, accurately segmenting cellular structures from electron microscopy data is crucial. To tackle this challenge, I recently trained a YOLO 11 detection model on the electron microscopy dataset provided by the Computer Vision Laboratory (CVLAB) at EPFL. This approach, combined with executing the model in LabVIEW using Graiphic’s deep learning toolkit, marks a significant breakthrough in terms of efficiency and accuracy.
Training YOLO 11 on Complex Data
The dataset used represents a 5x5x5 µm section of the brain’s hippocampus, capturing details at a resolution of 5x5x5 nm per voxel. The training focused on segmenting mitochondria, a key cellular structure. With YOLO 11’s enhanced architecture, I optimized feature extraction and improved detection precision while reducing inference time.
The training employed advanced data augmentation techniques and regularization strategies to enhance the model’s robustness against image variations. YOLO 11’s new adaptability and efficiency proved particularly effective for identifying and segmenting complex structures in such a detailed and rich environment.
Integration with LabVIEW and Graiphic’s Deep Learning Toolkit
The real innovation in this project lies in integrating the YOLO 11 model within the LabVIEW environment, using Graiphic’s deep learning toolkit. This integration allows researchers and engineers to combine the power of advanced image analysis with LabVIEW’s intuitive interface, streamlining deployment and result analysis.
Running the model in LabVIEW enabled real-time prediction visualization and leveraged the platform’s rapid prototyping and data acquisition capabilities. Graiphic’s infrastructure, which is built on ONNX runtime, optimized inference handling and accelerated workflow.
Results and Prospects
Testing demonstrated faster and more accurate segmentation of mitochondria, even in high-noise images. This approach of using YOLO 11 in LabVIEW opens up new possibilities for processing electron microscopy images, offering significant productivity gains for research teams and laboratories.
The unique combination of YOLO 11 and Graiphic’s deep learning toolkit holds the potential to revolutionize automated analysis in biomedical imaging, facilitating anomaly detection and the understanding of complex cellular structures.
Integration into LabVIEW
With the LabVIEW deep learning module, LabVIEW users will soon be able to benefit from advanced features to run models like YOLO V11. This module will enable these models to operate on LabVIEW, providing top-tier performance for complex computer vision tasks. Key features offered by this toolkit include:
- Full compatibility with existing frameworks: Keras, TensorFlow, PyTorch, ONNX.
- Impressive performance: The new LabVIEW deep learning tools will allow executions 50 times faster than the previous generation (HAIBAL 1.0) and 20% faster than PyTorch.
- Extended hardware support: CUDA, TensorRT for NVIDIA, Rocm for AMD, OneAPI for Intel.
- Maximum modularity: Define your own layers and loss functions.
- Graph neural networks: Complete and advanced integration.
- Annotation tools: An annotator as efficient as Roboflow, integrated into our software suite.
- Model visualization: Utilizing Netron for graphical model summaries.
- Generative AI: Complete library for execution, fine-tuning, and RAG setup for Llama 3, Phi 3, Florence-2 models.
Example Video
By integrating the advanced analysis of YOLO 11 with LabVIEW’s capabilities, large-scale processing of electron microscopy data has become more efficient. This solution paves the way for more diverse applications in neuroscience, bioinformatics, and other fields that require precise analysis of high-resolution images.